We really enjoyed meeting our members again at the symposium, and are very happy that we have welcomed a group of new members! Looking forward to meeting you after the holidays!

We really enjoyed meeting our members again at the symposium, and are very happy that we have welcomed a group of new members! Looking forward to meeting you after the holidays!

Former chair of the NVK, Dr. Raimond Ravellli, will provide his inaugural lecture to be appointed Professor in Structural Biology on Friday November 18th, 2022 at Maastricht University. The title of this lecture is “Musing on Nanobiology” and will also be broadcasted through a live-stream. Please find more information here.
On his inauguration, Raimond Ravelli showed a beautiful animation of the SARS-CoV-2 virus life cycle, watch it here.
We are looking for someone with affinity for social media, who is willing to contribute in keeping track of the Twitter and LinkedIn accounts of the NVK. Please contact us if you are interested, it would be of great help!

We (Sven Hennig and his team) are very excited to be part of Tom Grossmann’s project of fighting colorectal cancer using peptidomimetics. Tom just received an ERC PoC grant which is accelerating the kick-off for this endeavor. Together we have identified a family of peptidomimetic agents that bind beta-catenin and inhibit its interaction with the TCF/LEF transcription factors . For the first time, it was possible to obtain a crystal structure of a synthetic molecule bound to a therapeutically very attractive site on beta-catenin. In addition, we have confirmed cellular activity of these inhibitors verifying selective inhibition of the Wnt signalling pathway (Wendt et al. Angewandte Chemie, 2021, https://doi.org/10.1002/anie.202102082). These findings provide the ideal starting point for the development of novel therapeutics for Wnt-dependent cancers, in particular for CRC. Press: https://www.eurekalert.org/news-releases/767299.
proCOLON: ERC Proof-of-Concept granted to investigate first-in-class beta-catenin inhibitors as treatment for colorectal cancer.

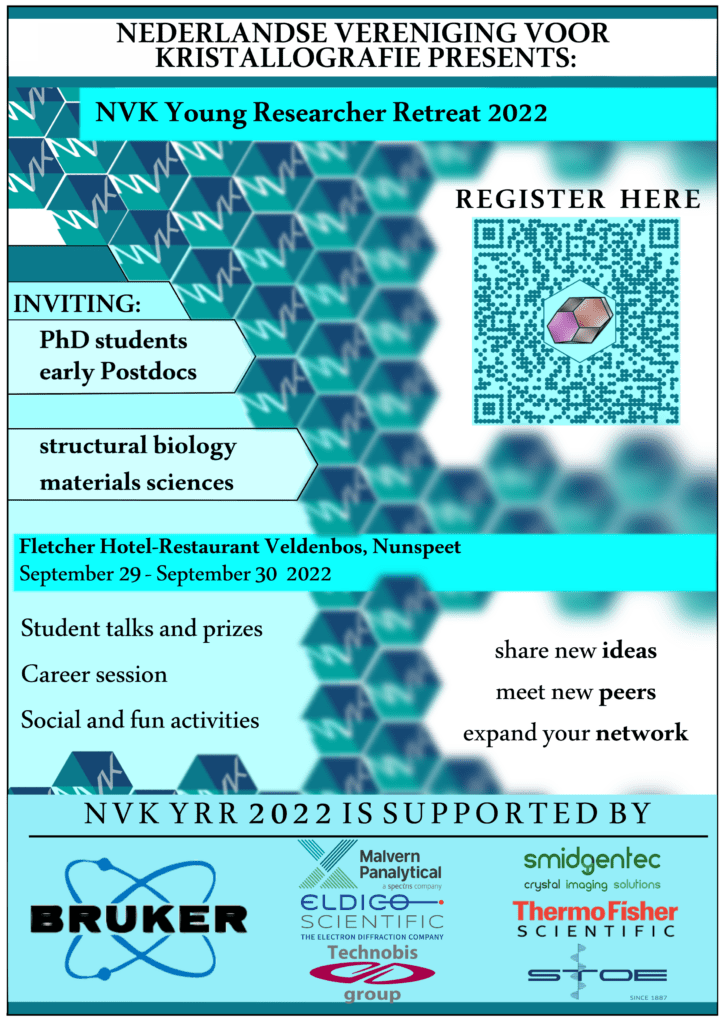
On September 29th and 30th, 2022, we are organizing the NVK Young Researchers Retreat! Please find the preliminary program and registration details here.
All participants will present their work, both PhD students and early researchers working in industry or industry are welcome to contribute, which will result in a wonderful and inspiring program! Our key-note speaker, Dr. Yesid A. Ramirez, will talk about his research related to . Additionally there will be plenty of time to network with our sponsors from industry, and of course with your fellow young crystallographers. Also, the career session will provide a overview of career options and plenty of time to discuss these with people who have been in the field for a longer time, both industry and academic science.
In short: interesting science, plenty of options to network, think a bit about your future career, and of course we will have a lot of fun! We hope to see you there!
I am working in crystallography for over 40 years. I came to our lab (Laboratorium voor Kristalchemie) in 1977 as a student and worked on solving carbohydrate crystal structures with Jan Kanters. We continued this work in my PhD (1980-1985, Prof. A.F Peerdeman), when I got also interested in modelling, both with quantum chemical methods and molecular mechanics/dynamics. After my PhD, I continued working on carbohydrates and in particular on cellulose, derivatives and wood fibers with my husband Jan Kroon (then the professor of our group), with whom I did many projects financed by industry (AkzoNobel), Economic Affairs (IOP-K and EET), EU-FAIR and STW (Fig. a&b). I was a post-doc during more than 15 years (impossible these days!), but I did not mind: we were always successful in getting grants for interesting projects and I was very happy to stay with Jan in his lab. That situation changed completely when Jan died unexpectedly in 2001. Piet Gros took over the lead of the group who already transformed it into a protein crystallography lab. I was appointed assistant professor, not without the help of the dean of chemistry, Prof. J.F.G Vliegenthart, in 2003. My research changed to methods developments in data processing, an area that I already became interested in because of my work on fiber diffraction. More recently, I became interested in diffuse scattering in both chemical and macromolecular crystallography (Fig. c). I have been active in the Diffraction Data Deposition Working Group (DDDWG) of the IUCr (International Union of Crystallography) from 2011 onward, and which now turned into a standing committee CommDat. We are setting up a new section in IUCrData: the Raw Data Letter, with the aim describing interesting, unusual, intriguing or complicated diffraction data, that could be a challenge for methods or software developers, but also for creating visibility of the data, which should be deposited in an archive with a persistent identifier (e.g. a DOI). More news on this will follow in future News Letters.

The period around 1972, 50 years ago was in hindsight a time of change. Much of the research lines of the crystallography group in Utrecht had been curiosity driven and were related to research started after WW-II by Professor Bijvoet. Among his many interests were the development of the isomorphous replacement techniques for solving crystal structures and the ground breaking use of anomalous dispersion to determine the absolute configuration of chiral molecules. The first such an absolute structure determination was done by Bijvoet and his PhD students Peerdeman (his successor) and van Bommel on a rubidium salt of tartaric acid using X-ray diffraction photographic film methods. This result, that was consistent with the arbitrary choice by Fisher, was first reported in 1949 at a meeting in the USA. This led to investigations of the crystal structures of other tartrates and similar small molecules such as malonic acid and the study of hydrogen bonding. Those were light atom compounds that needed different methods to solve the phase problem as those available for heavy atom structures. This spawned local interest in and development of so called Direct Methods, in particular of a technique called Symbolic Addition that worked well for solving centro-symmetric structures. X-ray diffraction data were collected with labour intensive film techniques and later on with semi-automatic three-circle single point detector based diffractometers for which in-house data collection software was developed. Computer based calculations and software development needed for the structure determination were done initially on a Dutch made in-house ZEBRA computer, being the first one at Utrecht University. That computing facility was, when I started in 1966 as a crystallography student, already replaced by a Dutch made Electrologica X8 computer at the university computer center elsewhere in the city. The latter was an Algol60 language, one job at a time, air-conditioned large room sized system with a computing power comparable to that of an early IBM PC. During daytime, small calculation jobs with data and program supplied on paper tape were run by computer operators at the computer center. Those tapes and the output results, again often in paper tape format and line printer output, were transported twice a day by a dedicated person, who also managed the Xerox copying machine, between the lab and the computing center. Turn-around time was often one day. A correction of input errors took another day. Larger jobs such as least-squares refinement were done mainly once a week during a 6 PM to 8 AM night shift where our crystallography group had the university computer for ourselves. Many users of the computer acted both as researcher, programmer and operator doing their calculations in turn. Coming back to the lab in the morning there was always coffee ready, provided by the housekeeping ladies to keep us awake that day. All that changed within a short period 50 years ago. The crystallography lab was housed in a free standing stately city house, also known as the ‘Crystal Palace’. The building was until the retirement of Bijvoet in 1962 in part laboratory and in part the house of the Bijvoet family. The lab moved in 1973 to a new building in the new University campus outside of the city. The computer center had moved by that time also to that campus area (de Uithof). A new multi-user American state-of-the-art Control Data computer system running Fortran had replaced the single user X8 by then. Paper tape supplied data and software were suddenly old-fashioned and practically unusable. The new standard was now IBM punch cards. All local software that was developed and used for years suddenly became obsolete and was replaced by an also American Fortran based crystallographic software package named XRAY72. Calculation jobs were now carried out in batch mode and could be submitted with an in-house terminal/lineprinter input/output station connected to the computer center. Small jobs could even be run interactively in time sharing mode. The group had now also obtained a new CAD4 diffractometer that potentially allowed for the collection of diffraction data for in the order of 25 structure studies per year. Structure determinations that took multiple months could now be completed within several weeks. One of the bottlenecks of a successful structure determination, the solution of the phase problem, was now also addressed significantly with the availability of the Direct Methods program MULTAN. Part of the research became also more collaborative, in particular with metal-organic and coordination chemistry groups. Research shifted in the direction of intermolecular interactions, structure prediction and probability based theory aiming to improve the success of the Direct Methods. It was interesting to be part of all those developments that allows us today, when pushed, to have in a routine case a structure report ready within a day starting with a good crystal. Unfortunately, much of the details of the procedures used will be hidden in black box software for the current crystal structure analist.
Written by Ton Spek
My name is Daan Swarts, and since 2019 I am an assistant professor at Wageningen University in the Laboratory of Biochemistry. In my independent research group, we focus on the characterization of bacterial immune systems. Also bacteria can get ‘sick’: invading nucleic acids such as viruses (bacteriophages) can kill cells, and other mobile genetic elements such as plasmids and transposons can be a metabolic burden or reduce fitness. Bacterial immune systems protect their host against such mobile genetic elements. Well-known examples of such systems are restriction-modification systems and CRISPR-Cas, but our research focuses on distinct immune systems of which the evolution, functionality, mechanisms, and structures are unknown. We are mainly interested in these systems from a fundamental point of view, but if interesting systems are found they might be repurposed for DNA editing or detection.
During my PhD (performed in the group of Prof. dr. John van der Oost at Wageningen University) my interest in bacterial immune systems was raised: I focused on the characterization of prokarytic Argonaute proteins (pAgos), which are homologous to eukaryotic Argonaute proteins (the key enzyme in RNA silencing pathways). We uncovered that pAgos interfere not with RNA but with DNA, and can protect their against invading DNA. During my PhD I got interested in protein structures, and wanted to learn how to apply structural biology methods. I continued my research as a post-doctoral EMBO fellow in the group of Prof. dr. Martin Jinek at the University of Zurich. There, my research revolved around determination of the structure and mechanisms of CRISPR-Cas12a, a CRISPR effector enzyme with capacities similar to that of Cas9. I learned how to apply X-ray crystallography and interpret structures, which remains important in the research we currently perform in my research group.

We investigate prokaryotic immune systems using research techniques from various fields including bacterial genetics and biochemistry. X-ray crystallography is used to determine the macromolecular structures of proteins in complex with the nucleic acids that they interact with. Combined, this allows us to chart in detail the function and biochemical mechanisms of uncharacterized prokaryotic immune systems. Funded by a VENI, an ERC starting grant, and some independent PhD grants, my team currently consists of six PhD students and a technician. We recently published the first research paper from my group, in which we characterized an Argonaute-based immune system that kills its host to prevent spread to other bacteria (sorry no structures (yet…)).
In my free time (as far as that exists next to the tenure track), I like to spend time with my family (my girlfriend and I have a daughter (4y) and a son (1y)), travel, be outside in nature, or play (board)games with friends. I also go to the gym and play field hockey to relieve pressure. During the corona pandemic, I also picked up gardening as a hobby. I am looking forward to meet researchers in the Dutch crystallography field in person, now this is possible again!
Here it is: the first online NVK newsletter! Please have a read and let us know what you think!
Also, please remember to register for the NVK Young Researchers Retreat.
Albert Guskov is a prominent structural biologist working on various membrane proteins (metal, vitamin, and amino acid transporters) and large molecular machineries (ribosomes and photosystems) using the combination of macromolecular crystallography, single particle cryo-electron microscopy and molecular dynamics simulations. He was trained in structural biology at the Institute of Protein Research of Russian Academy of Sciences (MSc, 2005) and in 2009 he received his PhD degree at Free University of Berlin under supervision of the renown crystallographer Prof Dr Wolfram Saenger. In 2010-2012 Albert stayed at the Nanyang Technological University in Singapore as a postdoc with Said Eshaghi, where he started working on membrane transporters and channels. In 2013, he moved to the Netherlands, where he built his career from a senior PostDoc to Associate Professor and currently he is the head of Biomolecular X-ray crystallography laboratory at the University of Groningen; from 2019 to 2022 he was also a visiting professor at the Moscow Institute of Physics and Technology, where he was the head of cryo-Electron Microscopy laboratory. Albert Guskov has published over 50 scientific publications (>3400 citations, h-index 25) and is regularly invited as a speaker to the national and international scientific meetings. Guskov’s lab at the University of Groningen combines state-of-the-art techniques of structural biology, namely macromolecular X-ray crystallography, single-particle cryo-electron microscopy and molecular dynamics simulations to tackle various important biological questions. The main research lines of the lab are as follows: